The function dhhh4sims constructs a (non-vectorized)
probability mass function from the result of
surveillance::simulate.hhh4() (and the corresponding
model), as a function of the time point within the simulation period.
The distribution at each time point is obtained as a mixture of
negative binomial (or Poisson) distributions based on the samples from
the previous time point.
dhhh4sims(sims, model)Arguments
- sims
a
"hhh4sims"object fromsurveillance::simulate.hhh4().- model
the "hhh4" object underlying
sims.
Value
a function(x, tp = 1, log = FALSE), which takes a
vector of model$nUnit counts and calculates the
(log-)probability of observing these counts (given the
model) at the tp'th time point of the simulation
period (index or character string matching rownames(sims)).
See also
logs_hhh4sims() where this function is used.
Examples
library("surveillance")
#> Loading required package: sp
#> Loading required package: xtable
#> This is surveillance 1.22.1; see ‘package?surveillance’ or
#> https://surveillance.R-Forge.R-project.org/ for an overview.
CHILI.sts <- sts(observed = CHILI,
epoch = as.integer(index(CHILI)), epochAsDate = TRUE)
## fit a simple hhh4 model
(f1 <- addSeason2formula(~ 1, period = 365.2425))
#> ~1 + sin(2 * pi * t/365.2425) + cos(2 * pi * t/365.2425)
fit <- hhh4(
stsObj = CHILI.sts,
control = list(ar = list(f = f1), end = list(f = f1), family = "NegBin1")
)
## simulate the last four weeks (only 200 runs, for speed)
sims <- simulate(fit, nsim = 200, seed = 1, subset = 884:nrow(CHILI.sts),
y.start = observed(CHILI.sts)[883,])
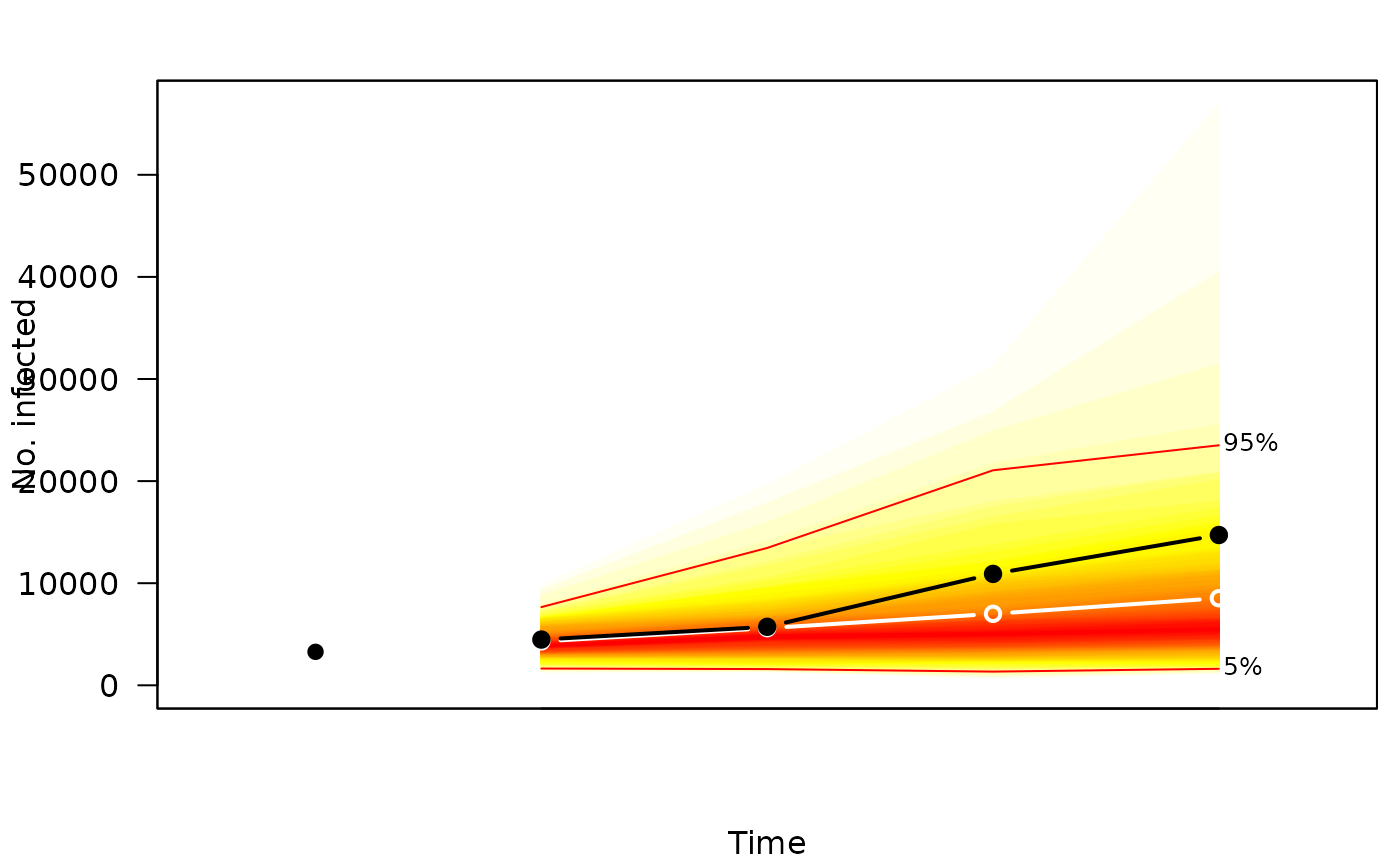
if (requireNamespace("fanplot")) {
plot(sims, "fan", fan.args = list(ln = c(5,95)/100),
observed.args = list(pch = 19), means.args = list(type = "b"))
}
#> Loading required namespace: fanplot
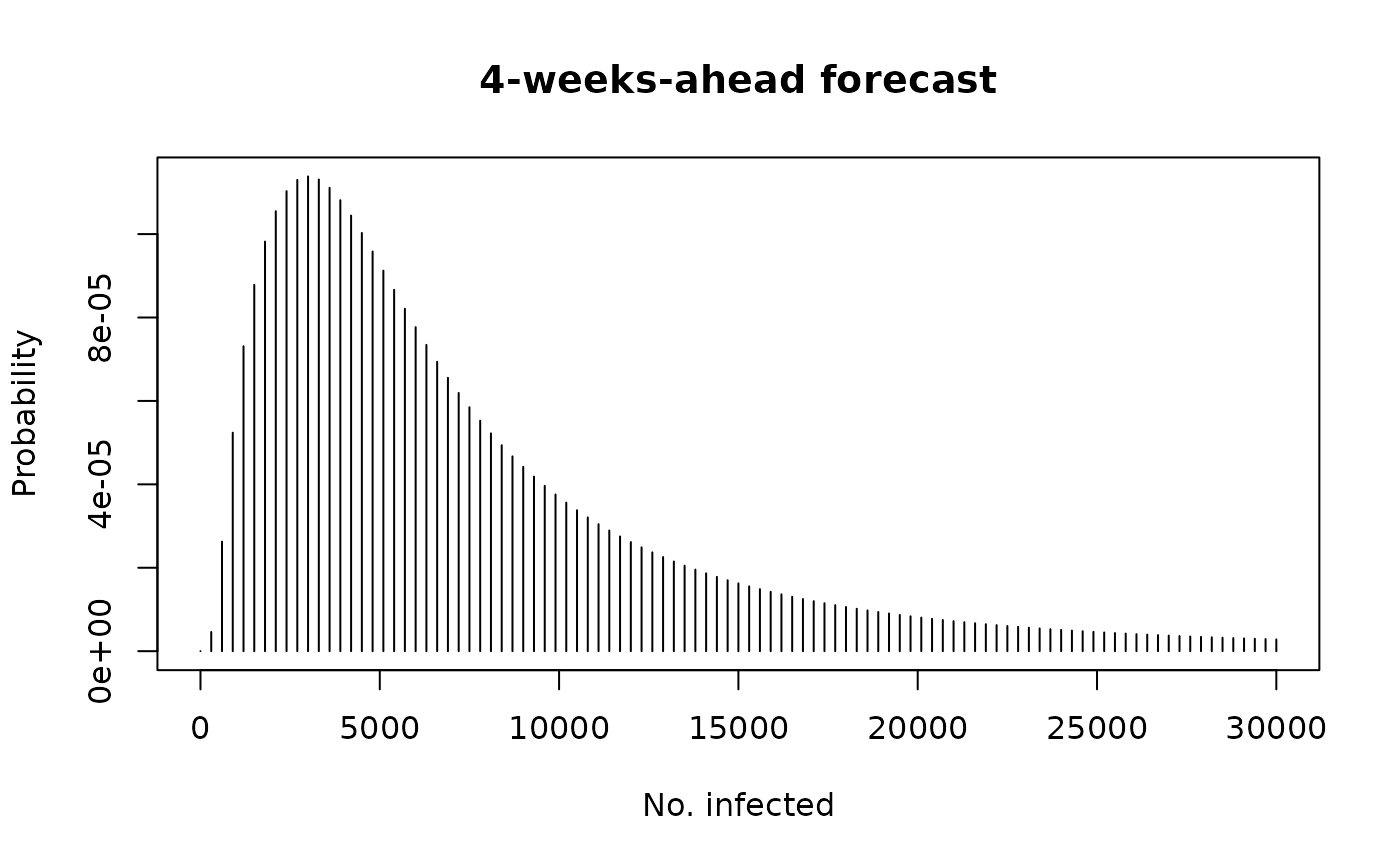
 ## derive the weekly forecast distributions
dfun <- dhhh4sims(sims, fit)
dfun(4000, tp = 1)
#> [1] 0.0002194578
dfun(4000, tp = 4)
#> [1] 0.0001069622
curve(sapply(x, dfun, tp = 4), 0, 30000, type = "h",
main = "4-weeks-ahead forecast",
xlab = "No. infected", ylab = "Probability")
## derive the weekly forecast distributions
dfun <- dhhh4sims(sims, fit)
dfun(4000, tp = 1)
#> [1] 0.0002194578
dfun(4000, tp = 4)
#> [1] 0.0001069622
curve(sapply(x, dfun, tp = 4), 0, 30000, type = "h",
main = "4-weeks-ahead forecast",
xlab = "No. infected", ylab = "Probability")
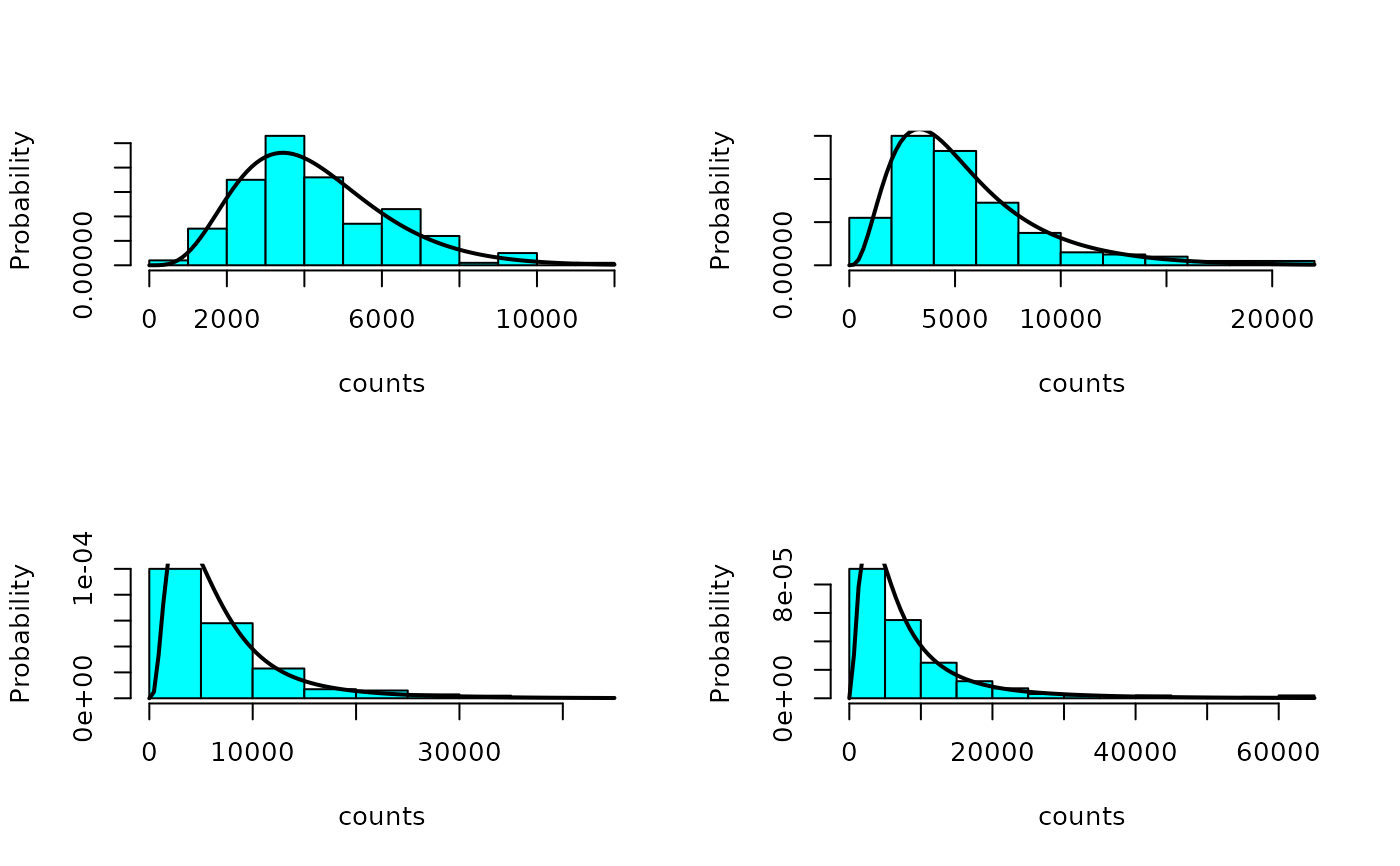
 ## compare the forecast distributions with the simulated counts
par(mfrow = n2mfrow(nrow(sims)))
for (tp in 1:nrow(sims)) {
MASS::truehist(sims[tp,,], xlab = "counts", ylab = "Probability")
curve(sapply(x, dfun, tp = tp), add = TRUE, lwd = 2)
}
## compare the forecast distributions with the simulated counts
par(mfrow = n2mfrow(nrow(sims)))
for (tp in 1:nrow(sims)) {
MASS::truehist(sims[tp,,], xlab = "counts", ylab = "Probability")
curve(sapply(x, dfun, tp = tp), add = TRUE, lwd = 2)
}
 # \dontshow{
## verify distribution at the first time point (i.e., one-step-ahead NegBin)
stopifnot(identical(
sapply(0:100, dfun, tp = 1),
dnbinom(0:100,
mu = meanHHH(fit$coefficients, terms(fit), subset = 884, total.only = TRUE),
size = sizeHHH(fit$coefficients, terms(fit), subset = 884))
))
## check that we have a probability distribution at the last time point
.xgrid <- seq(0, 100000, by = 500)
stopifnot(abs(1 -
integrate(approxfun(.xgrid, sapply(.xgrid, dfun, tp = 4)), 0, 100000)$value
) < 0.001)
# }
# \dontshow{
## verify distribution at the first time point (i.e., one-step-ahead NegBin)
stopifnot(identical(
sapply(0:100, dfun, tp = 1),
dnbinom(0:100,
mu = meanHHH(fit$coefficients, terms(fit), subset = 884, total.only = TRUE),
size = sizeHHH(fit$coefficients, terms(fit), subset = 884))
))
## check that we have a probability distribution at the last time point
.xgrid <- seq(0, 100000, by = 500)
stopifnot(abs(1 -
integrate(approxfun(.xgrid, sapply(.xgrid, dfun, tp = 4)), 0, 100000)$value
) < 0.001)
# }